EVLncRNAs User Tutorial
The usage of the EVLncRNAs database is as follows:
1. In ‘Browse’ menu, there are four pull-down menus: Species, Disease, Interaction and Peptide-related, and each has submenus. Users can browse relevant entries in EVLncRNAs by clicking any pull-down menu or their submenus and the page will move to the related page. Then users also can click navigation bar on the left to view other pages or corresponding entries (Figure 1).
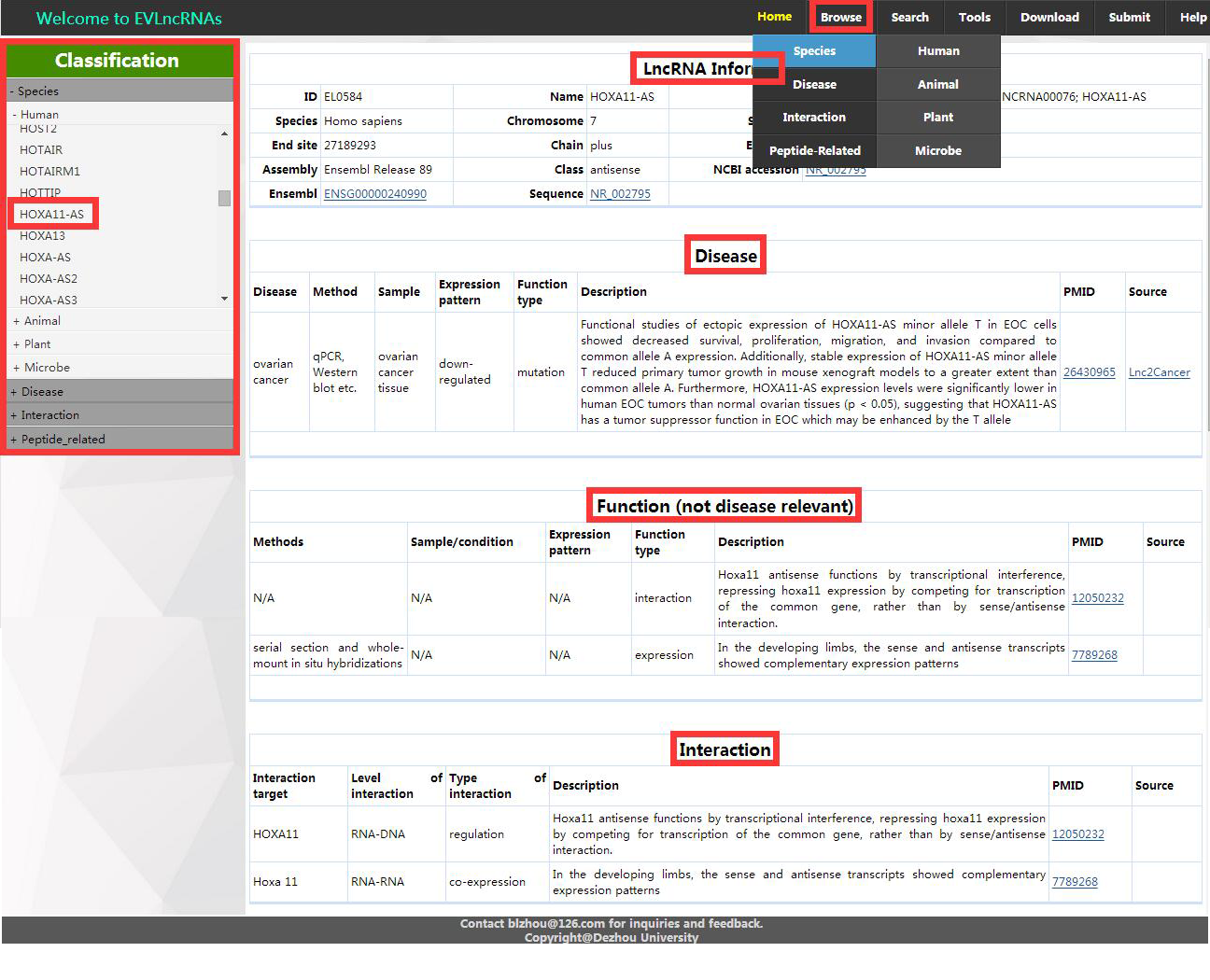
Figure 1. The Browse page of the EVLncRNAs
a) Each entry in ‘Species’ category has the following items (The blank part is not shown):
-
LncRNA Information:
ID, Name, Aliases,
Species,Chromosome,
Start site, End site, Chain,Extron NO.,
Type, NCBI accession, Ensembl
-
Disease:
Disease, Method,
Sample, Expression
pattern, function
type, Description, PMID, Source
-
Function:
Method, Sample/Condition,
Expression pattern,
function type, Description, PMID, Source
-
Interaction:
Interaction target,
Level of interaction,
Type of interaction.
Description, PMID, Source
function type: According to the experimentally supported associations between lncRNAs and diseases, the function types were classified into expression, mutation, interaction, locus, and epigenetics.
Level of interaction: levels of interaction components, for example, RNA-DNA, RNA-RNA, and RNA-Protein. Type of interaction: According to the way how the molecules interact, types of interaction were classified into binding, regulation and co-expression.
b) Each entry in ‘Disease’ category has the following items:
-
Disease
LncRNA ID, lncRNA
Name, Disease,
Method, Sample,
Expression pattern, function type,
Description, PMID, Source
c) Each entry in ‘Interaction’ category has the following items:
-
Interaction:
LncRNA ID, lncRNA
Name, Interaction
target, Level of
interaction, Type of interaction.
Description, PMID, Source
d) Each entry in ‘Peptide-related’ category has the items same as those in ‘Species’ category.
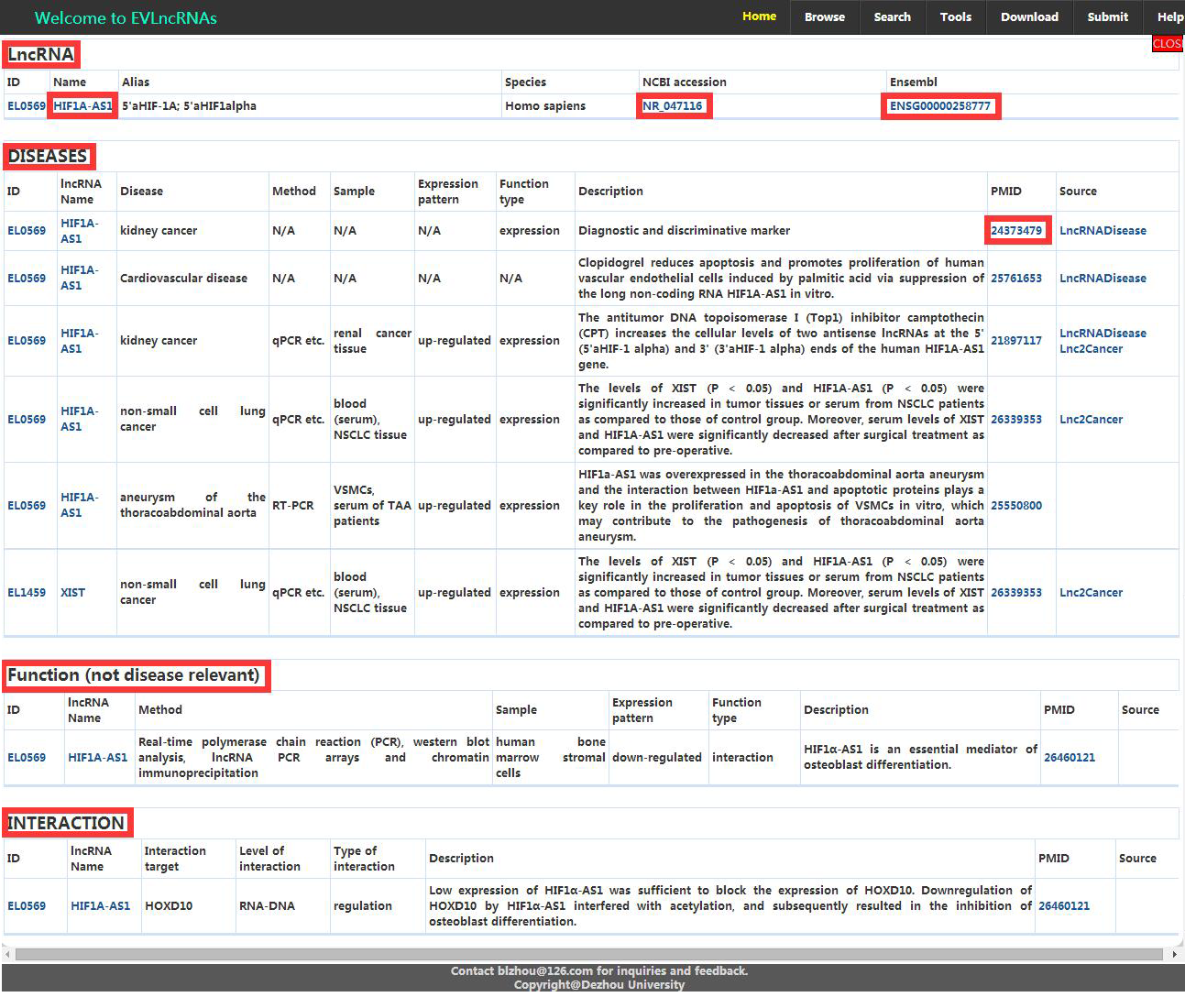
2. In the ‘Search’ page, EVLncRNAs enables users to search by any items, such as, lncRNA name, alias, disease name, experimental methods, associated components, level of interaction and so on. EVLncRNAs offers ‘fuzzy’ searching capabilities and returns all matching records. The search results are linked to the pages of related lncRNAs and original articles (Figure 2).
3. EVLncRNAs provides reliable and available tools for lncRNA prediction and lncRNA function prediction in the ‘Tools’ page.
4. EVLncRNAs supports users to download the database data in the ‘Download’ page.
5. EVLncRNAs invites users to submit novel experimentally validated lncRNAs and related diseases or associated components in ‘Submit’ page. The submitted record approved by our researchers will be included in the following release.
If there are any comments or suggestions, please contact at blzhou@126.com